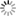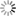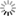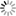Jack Lucentini, "Is This Life?" (2006)
"The Scientist" January 2006 Volume 20 | Issue 1 | Page 30; http://www.the-scientist.com/2006/1/1/30/1/
|
|||
 |
|||
|
HORDES OF GREEN, SUB-MICROSCOPIC BALLOONS FLOAT in a watery mixture in Jack Szostak's laboratory at Harvard Medical School. They come in a variety of shapes: spheres, blimps, worms. And as Szostak examines magnified images of them, he can't help but notice a striking resemblance to bacterial ecosystems, pulsing with that fetid, yet undeniable quality that has eluded definition for generations - life. But these orbs aren't alive. The uncanny resemblance reflects the fact that these ersatz sacs may passably mimic the wrappings of primitive life: cell membranes. But infusing in them the real "stuff" of life requires more work. Lately, Szostak, a professor of genetics, has been putting simple RNA enzymes inside, showing that they can conduct their characteristic activities. Thus some of life's chemistry is compatible with artificial membranes, he says, something that required a careful tweaking of the membrane chemistry. He has also made the sacs grow spontaneously, and even divide - with help.1 "It's a simplified model of the situation we'd really like to have," says Szostak: a growing, dividing, living organism of totally synthetic origins. But even at present, he says, "These simple membrane systems do pretty fascinating things." 12 STEPS TO A NEW LIFE
Ever since chemist Stanley Miller created organic compounds from simple building blocks like water, methane, and ammonia, the idea of creating life and thus peering into its possible origins, has fascinated biologists. David W. Deamer, professor emeritus of chemistry and biochemistry at the University of California, Santa Cruz, and a cadre of pioneers expanded the quest three decades ago, launching an attempt to build a "protocell." According to Deamer, such an entity must meet 12 requirements for life including having membrane enclosures (1) that can capture energy (2), maintain ion gradients (3), encapsulate macromolecules (4), and divide (5). Macromolecules must be able to grow by polymerization (6), evolve in a way that speeds growth (7), and store information (8). Add to that information store the ability to mutate (9) and to direct growth of catalytic polymers, and you have 10.2 In the past decade, Deamer says, individual labs have met each of these requirements but in quite different ways (see The Final Step(s)? for more discussion). With only two steps remaining, they might achieve a synthetic organism within this decade. Many of the people approaching this are engineers, sharing in the philosophy that one can't truly understand what one can't build. Albert Libchaber of Rockefeller University engineered a DNA plasmid to express proteins and put them into membranous sacs. They could produce proteins for a few hours but would eventually peter out when the raw materials ran low inside the compartment. They needed to keep the supply coming. So, he and Vincent Noireaux, now an assistant professor at the University of Minnesota, designed them to produce a channel-forming protein, alpha hemolysin.3 Suddenly, finished proteins tagged with Green Fluorescent Protein inserted themselves into the artificial membrane allowing nucleotides and other molecules to enter. These "cells" survive for up to four days, but it's only a small victory. In the quest to build life, defining success is hard, Libchaber says. Is it success simply to create a cell that functions? Or must it also reproduce? "I think in our case at least, the first step has been achieved." Next, he wants to make them divide, something that's only been done thus far through physical manipulation. Such is the focus of the remaining two steps, says Deamer. The cell must contain genes and enzymes that can be replicated (11), and they must be shared among daughter cells (12). "We don't have a way to make the entire system of catalysts, all growing together," that drives cellular reproduction, he explains. And, he adds, the final stretch may boil down to a single achievement. An enzyme that duplicates itself, such as a self-replicating ribozyme, might do the job, acting as both genetic material and the catalyst for replication. Roughly a dozen laboratories worldwide are working on it, Deamer says, and this has spurred major progress. DEFINING SQUARE ONE
Progress has not simply been made through a single approach, of course. Others have been attempting to create artificial life by overhauling genetic instructions. © COURTESY OF MARTIN HANCZYC

THE GLOWING, GROWING HORDES:
> RNA, with red fluorescent dye adsorbs to the surface of a montmorillonite clay particle encapsulated by a fatty acid vesicle labeled with green fluorescence. This structure forms through a process of self organization mediated by the clay, and illustrates a possible pathway by which the precursors of the first living cells could have formed.<div> For years, scientists have been approaching genomes and looking to strip out nonessential material. The J. Craig Venter Institute worked extensively with the bacterium Mycoplasma genitalium, using comparative genomics and mutation analyses to identify an estimated 265 to 350 core genes required for life. Hamilton O. Smith and collaborators plan to synthesize an entire artificial chromosome based on this "minimal genome," and insert it in pieces into a living cell. Using recombinational mechanisms from Deinococcus radiodurans (which famously reconstructs its entire genome after disruption by ionizing radiation) they will attempt to rebuild it, says Smith. Such so-called top-down approaches start with something that already works, says Mark Bedau, professor of philosophy and humanities at Reed College in Portland, Ore., and editor-in-chief of the journal Artificial Life. But bottom-up approaches such as those by Szostak and Libchaber may reveal "much more about what's possible," he adds, "because you're making everything from scratch." And starting from scratch provides significant opportunities, says Bedau: "You're not constrained by the historical accidents found in the existing forms of life." That could lead to life forms more like the first life thought to be on Earth, or even like life on other planets. Deamer says it will take a wide range of approaches, both top-down and bottom-up, to reach the goal that unites most researchers in the field: the drive to learn "what the scaffolding was that let this kind of a cell crawl out of the primordial ooze." Liaohai Chen, a molecular biologist at Argonne National Laboratory in Illinois, says that precisely because such a wide range of strategies is underway, it's not so easy to generalize about how far "protocell" research has gotten, nor what the final hurdle will be. "The hurdle is really project dependent," he says. Indeed, in attempts to create artificial life Chen along with Steen Rasmussen of Los Alamos National Laboratory have thrown out much of the conventions found in nature. They've turned the protocell model inside out, designing a micelle with information coding and metabolic machinery on its exterior.4 Extant thus far mostly on paper, these micelles use peptide nucleic acids (PNAs), DNA-mimics with a pseudopeptide backbone conjugated to a light-sensitive molecule. When exposed to light, the photosensitive chemical discharges an electron triggering chemical reactions to convert nearby nutrients into new fatty acids and PNA based on the PNA template. These incorporate into the micelle, which grows until it spontaneously pinches in half and divides. "Cell production is the area where would-be protocell-designers have made the least progress."
> <div style="MARGIN-RIGHT: 150px" align="right"><em>-Pier Luigi Luisi<em> For now, these inventions exist mainly as computer simulations; real-life testing is only just beginning. Rasmussen says the design's main advantage is its simplicity, which increases the chances it will work. "[It's] probably not how the origin of life historically happened, but it's the simplest thing we could come up with," he adds. "We're trying to understand why and how matter can self-organize? and become living. What is it in nature that enables it to do that? That's the deep question here." Deamer calls it one of the most ambitious protocell designs: "[They're the] one group that now is devoted to a true attempt to make a growing, reproducing cellular system." The team has worked out a whole life cycle for its organisms, at least in simulation, and has a $5-million grant from Los Alamos for the project. But taking stricter lessons from nature, Tetsuya Yomo of the University of Osaka, Japan and colleagues created a membrane-bound system exhibiting not only gene expression, but a two-stage genetic cascade of events, in an attempt to capture some of the complexity of real cells.5 Liposomes loaded with a bacterial plasmid and SP6 RNA polymerase drive the production of a T7 RNA polymerase, which is needed to catalyze the transcription of a gene for the mutant green fluorescent protein under the control of a T7 promoter. Surprisingly, the system worked even without the addition of SP6. Such work mines the divide between top-down and bottom-up approaches. In many projects, says Bedau, "the design is determined from the top down, but the construction is still from the bottom up." CONQUERING DIVISION
Libchaber's plans to make a protocell reproduce borrow from, which he says requires only five genes to divide successfully. One of these, FtsZ, makes a protein that polymerizes to form the initial ring at mid-cell that pinches the membrane during cytokinesis. So far, Libchaber says, his team has succeeded in getting the protein to polymerize on the membrane. The next step will be "to play with combinations of phospholipid, so we can destabilize the membrane," he says; this would allow the compartment to divide more easily. Cell reproduction is the area where would-be protocell-designers have made the least progress, says Pier Luigi Luisi, a professor of biochemistry at the University of Rome 3. Bedau says a self-replicating enzyme would help with this problem immensely. But on that front, Bedau laments, "There hasn't been any real progress for a number of years, and not for lack of trying." In 2001, researchers used a ribozyme to catalyze not its own replication, but that of another RNA template molecule.6 In principle this kind of activity would be good enough, Deamer says, "as long as it made another ribozyme that could continue this process." But the molecule can only copy a 14-nucleotide sequence and is itself hundreds of nucleotides long. Looking at biology from the top-down might provide the information needed to create a minimal cell that meets Deamer's strict set of requirements. "Once you determine what the minimal gene set is, then you can possibly go ahead and try to find out whether you can make minimal cells," reasons Dusko Ehrlich, research director at the National Institute for Agricultural Research in France. Ehrlich led a team of researchers in a study to identify the minimum set of genes in Bacillus subtilis.7 The group narrowed the number down to 271 (see Hot Paper story). One "encouraging" finding, he adds, is that only 4% of the genes he identified had unknown functions, showing "we do know quite a bit about a cell." Luisi says he's following such studies closely, and will possibly use the results as a guide to build cells from scratch. But 271 is still far too many genes for his liking. By infusing droplets of commercially sold protein-expression solutions into liposomes, he has found that some cellular functions can occur with a mere 80 genes or so. Reducing the system below 15 components might lead to something that could be made from the ground up, he proposes. "The earliest cells may have contained maybe 10-15 components," he says. "Of course they were limping. That's the challenge of the research," he adds - to rediscover "these limping old-timers." References
1. M.M. Hanczyc et al., "Experimental models of primitive cellular compartments encapsulation, growth and division," Science, 302:618-22, 2003.
3. V. Noireaux, A. Libchaber, "A vesicle bioreactor as a step toward an artificial cell assembly," Proc Natl Acad Sci, 101:17669-74, 2004.
5. K. Ishikawa et al., "Expression of a cascading genetic network within liposomes," FEBS Lett, 576:387-90, 2004.
|